sklearn_housing_bunch = fetch_california_housing("~/data/sklearn_datasets/")
Downloading Cal. housing from https://ndownloader.figshare.com/files/5976036 to /home/brunhilde/data/sklearn_datasets/
print(sklearn_housing_bunch.DESCR)
California housing dataset.
The original database is available from StatLib
http://lib.stat.cmu.edu/datasets/
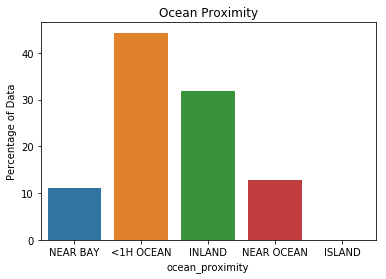
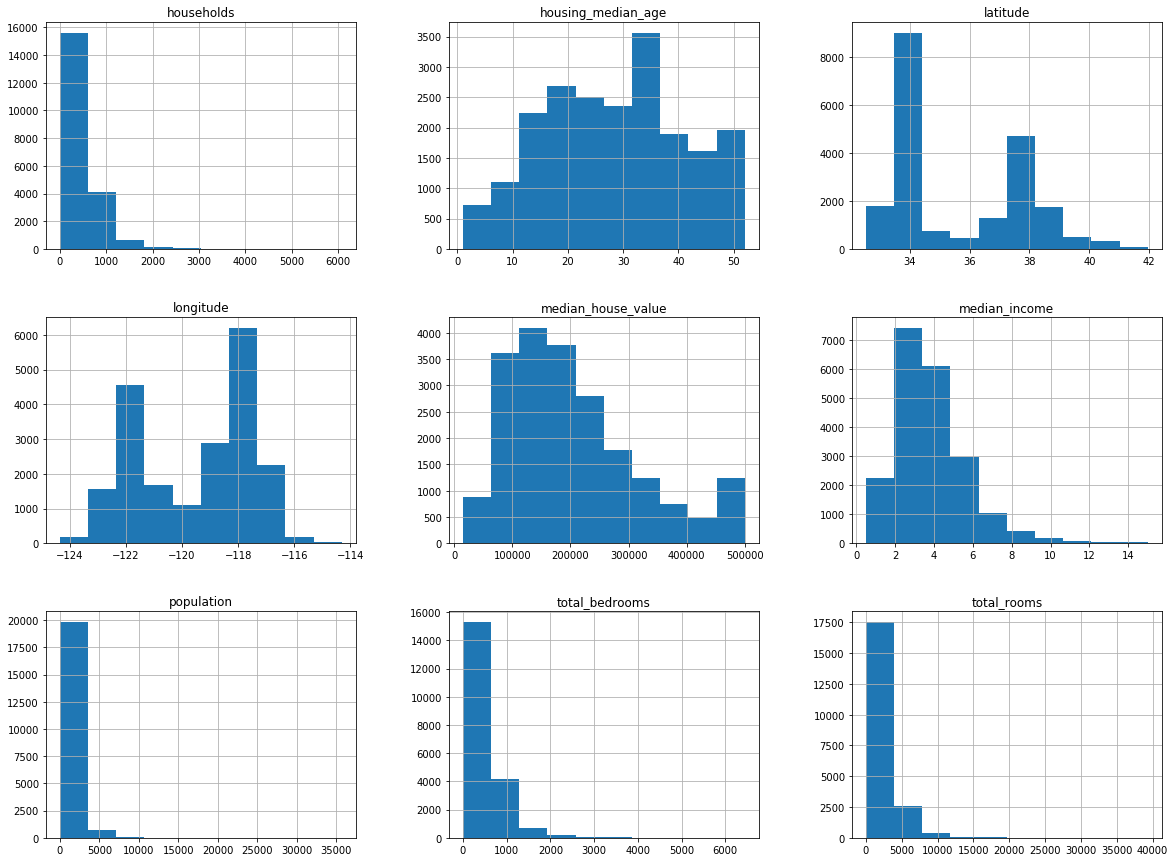
The data contains 20,640 observations on 9 variables.
This dataset contains the average house value as target variable
and the following input variables (features): average income,
housing average age, average rooms, average bedrooms, population,
average occupation, latitude, and longitude in that order.
References
----------
Pace, R. Kelley and Ronald Barry, Sparse Spatial Autoregressions,
Statistics and Probability Letters, 33 (1997) 291-297.
print(sklearn_housing_bunch.feature_names)
['MedInc', 'HouseAge', 'AveRooms', 'AveBedrms', 'Population', 'AveOccup', 'Latitude', 'Longitude']
Now I'll convert it to a Pandas DataFrame.
sklearn_housing = pandas.DataFrame(sklearn_housing_bunch.data,
columns=sklearn_housing_bunch.feature_names)
MedInc HouseAge AveRooms AveBedrms Population \
count 20640.000000 20640.000000 20640.000000 20640.000000 20640.000000
mean 3.870671 28.639486 5.429000 1.096675 1425.476744
std 1.899822 12.585558 2.474173 0.473911 1132.462122
min 0.499900 1.000000 0.846154 0.333333 3.000000
25% 2.563400 18.000000 4.440716 1.006079 787.000000
50% 3.534800 29.000000 5.229129 1.048780 1166.000000
75% 4.743250 37.000000 6.052381 1.099526 1725.000000
max 15.000100 52.000000 141.909091 34.066667 35682.000000
AveOccup Latitude Longitude
count 20640.000000 20640.000000 20640.000000
mean 3.070655 35.631861 -119.569704
std 10.386050 2.135952 2.003532
min 0.692308 32.540000 -124.350000
25% 2.429741 33.930000 -121.800000
50% 2.818116 34.260000 -118.490000
75% 3.282261 37.710000 -118.010000
max 1243.333333 41.950000 -114.310000