KNN Classification
1 Introduction
This looks at the performance of the K-Nearest Neighbors for classification. K-Nearest Neighbors works by finding the k (count) of neighbors that are closest to the data-point and classifying the point using the majority vote of those points. I'm going to use the default distance measurement of Euclidean distance. Fitting in this case means memorizing all the data so you can use it for predictions and then doing the calculations when you need to make a prediction. This makes it memory-intensive and slower when it's used to make predictions, so it's useful as a baseline, but not in production.
2 Synthetic
I'll start with a synthetic data set created by sklean. I'll make it the same shape as the Breast Cancer case that I'll look at later.
2.1 Imports
from sklearn.datasets import make_classification from sklearn.model_selection import train_test_split from sklearn.neighbors import KNeighborsClassifier import matplotlib.pyplot as pyplot import seaborn
%matplotlib inline seaborn.set_style("whitegrid")
2.2 The Data
total = 569 positive_fraction = 212/total negative_fraction = 1 - positive_fraction inputs, classifications = make_classification(n_samples=total, n_features=30, weights=[positive_fraction, negative_fraction]) print(inputs.shape) print(classifications.shape)
(569, 30) (569,)
positive = classifications.sum() print("Positives: {}".format(positive)) print("Negatives: {}".format(classifications.size - positive))
Positives: 355 Negatives: 214
X_train, X_test, y_train, y_test = train_test_split(inputs, classifications)
2.3 The model
model = KNeighborsClassifier()
def get_accuracies(max_neighbors=10): train_accuracies = [] test_accuracies = [] for neighbors in range(1, max_neighbors+1): classifier = KNeighborsClassifier(n_neighbors=neighbors) classifier.fit(X_train, y_train) train_accuracies.append(classifier.score(X_train, y_train)) test_accuracies.append(classifier.score(X_test, y_test)) return train_accuracies, test_accuracies
training_accuracies, testing_accuracies = get_accuracies()
neighbors = range(1, 11) pyplot.plot(neighbors, training_accuracies, label="Training Accuracy") pyplot.plot(neighbors, testing_accuracies, label="Testing Accuracy") pyplot.ylabel("Accuracy") pyplot.xlabel("Neighbors") pyplot.title("KNN Cancer Accuracy") pyplot.legend()
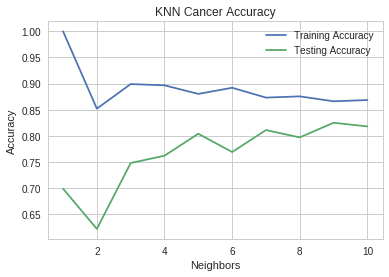
At k=1, the training set does perfectly while the test set does okay, but not as well as it does at k=9, what appears to be the best value.
3 Breast Cancer
3.1 Imports
import matplotlib.pyplot as pyplot import seaborn import pandas from sklearn.model_selection import train_test_split from sklearn.datasets import load_breast_cancer from sklearn.neighbors import KNeighborsClassifier
%matplotlib inline seaborn.set_style("whitegrid")
3.2 The Dataset
cancer = load_breast_cancer() print("Keys in the cancer bunch: {}".format(",".join(cancer.keys()))) print("Training Data Shape: {}".format(cancer.data.shape)) print("Target Names: {}".format(','.join(cancer.target_names)))
Keys in the cancer bunch: feature_names,target_names,target,data,DESCR Training Data Shape: (569, 30) Target Names: malignant,benign
This is from the description.
- Data Set Characteristics:
Number of Instances: 569
| Number of Attributes: | |
|---|---|
| 30 numeric, predictive attributes and the class | |
| Attribute Information: | |
- radius (mean of distances from center to points on the perimeter)
- texture (standard deviation of gray-scale values)
- perimeter
- area
- smoothness (local variation in radius lengths)
- compactness (perimeter^2 / area - 1.0)
- concavity (severity of concave portions of the contour)
- concave points (number of concave portions of the contour)
- symmetry
- fractal dimension ("coastline approximation" - 1)
The mean, standard error, and "worst" or largest (mean of the three largest values) of these features were computed for each image, resulting in 30 features. For instance, field 3 is Mean Radius, field 13 is Radius SE, field 23 is Worst Radius.
- class:
- WDBC-Malignant
- WDBC-Benign
| Missing Attribute Values: | |
|---|---|
| None | |
| Class Distribution: | |
| 212 - Malignant, 357 - Benign | |
| Creator: | Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian |
| Donor: | Nick Street |
| Date: | November, 1995 |
This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets. https://goo.gl/U2Uwz2
Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image.
Separating plane described above was obtained using Multisurface Method-Tree (MSM-T) [K. P. Bennett, "Decision Tree Construction Via Linear Programming." Proceedings of the 4th Midwest Artificial Intelligence and Cognitive Science Society, pp. 97-101, 1992], a classification method which uses linear programming to construct a decision tree. Relevant features were selected using an exhaustive search in the space of 1-4 features and 1-3 separating planes.
The actual linear program used to obtain the separating plane in the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: "Robust Linear Programming Discrimination of Two Linearly Inseparable Sets", Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server:
ftp ftp.cs.wisc.edu cd math-prog/cpo-dataset/machine-learn/WDBC/
References
- W.N. Street, W.H. Wolberg and O.L. Mangasarian. Nuclear feature extraction for breast tumor diagnosis. IS&T/SPIE 1993 International Symposium on Electronic Imaging: Science and Technology, volume 1905, pages 861-870, San Jose, CA, 1993.
- O.L. Mangasarian, W.N. Street and W.H. Wolberg. Breast cancer diagnosis and prognosis via linear programming. Operations Research, 43(4), pages 570-577, July-August 1995.
- W.H. Wolberg, W.N. Street, and O.L. Mangasarian. Machine learning techniques to diagnose breast cancer from fine-needle aspirates. Cancer Letters 77 (1994) 163-171.
Loading the Data
target = pandas.DataFrame(dict(target=cancer.target)) target_map = dict(zip(range(len(cancer.target_names)), cancer.target_names)) target['name'] = target.target.apply(lambda entry: target_map[entry]) print(target.name.value_counts())
benign 357 malignant 212 Name: name, dtype: int64
3.3 Splitting the Data
X_train, X_test, y_train, y_test = train_test_split(cancer.data, cancer.target, stratify=cancer.target) print("Trainining percent: {0:.2f} %".format(100 * len(y_train)/len(cancer.target))) print("Testing percent: {0:.2f}".format(100 * len(y_test)/len(cancer.target)))
Trainining percent: 74.87 % Testing percent: 25.13
3.4 Model Performance
def get_accuracies(max_neighbors=10): train_accuracies = [] test_accuracies = [] for neighbors in range(1, max_neighbors+1): classifier = KNeighborsClassifier(n_neighbors=neighbors) classifier.fit(X_train, y_train) train_accuracies.append(classifier.score(X_train, y_train)) test_accuracies.append(classifier.score(X_test, y_test)) return train_accuracies, test_accuracies
training_accuracies, testing_accuracies = get_accuracies()
neighbors = range(1, 11) pyplot.plot(neighbors, training_accuracies, label="Training Accuracy") pyplot.plot(neighbors, testing_accuracies, label="Testing Accuracy") pyplot.ylabel("Accuracy") pyplot.xlabel("Neighbors") pyplot.title("KNN Cancer Accuracy") pyplot.legend()
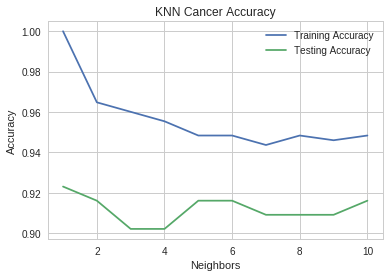
It looks like five neighbors would be what you'd want.
print("Minimum test accuracy (n=1): {:.2f}".format(min(testing_accuracies))) print("Maximum test accuracy (n=5): {:.2f}".format(max(testing_accuracies))) assert max(testing_accuracies == testing_accuracies[4])
Minimum test accuracy (n=1): 0.90 Maximum test accuracy (n=5): 0.92
The original paper that used this data-set got a cross-validation error-rate of 3%, but it sounds like they didn't split the data into training and testing sets (I'll have to re-read the paper to be sure).