Autocorrect: Minimum Edit Distance Backtrace
Beginning
This is the last post in a series about Autocorrect that's started in this post. In the previous post we implemented some code to find the minimum edit distance between two strings using Dynamic Programming. Now we'll implement the Backtrace Algorithim to find the shortest path through to transform one string to another.
Imports
# python
from argparse import Namespace
from functools import partial
from string import ascii_uppercase
import random
# pypi
from nltk.metrics.distance import edit_distance_align
from tabulate import tabulate
import holoviews
import hvplot.pandas
import matplotlib.pyplot as pyplot
import numpy
import pandas
import seaborn
# this repository
from neurotic.nlp.autocorrect.distance import MinimumEdits
# my other stuff
from graeae import EmbedHoloviews
Set Up
Plotting
This sets up some convenience code for plotting.
SLUG = "autocorrect-minimum-edit-distance-backtrace"
FOLDER = f"files/posts/nlp/{SLUG}/"
Embed = partial(EmbedHoloviews, folder_path=FOLDER)
Plot = Namespace(
width=990,
height=780,
fontscale=2,
tan="#ddb377",
blue="#4687b7",
red="#ce7b6d",
color_map="Plasma",
path_color_map="blues",
)
get_ipython().run_line_magic('matplotlib', 'inline')
get_ipython().run_line_magic('config', "InlineBackend.figure_format = 'retina'")
seaborn.set(style="whitegrid",
rc={"axes.grid": False,
"font.family": ["sans-serif"],
"font.sans-serif": ["Open Sans", "Latin Modern Sans", "Lato"],
"figure.figsize": (8, 6)},
font_scale=1)
FIGUE_SIZE = (12, 10)
Tabulate
This sets up a pretty-printer for path-tables.
PATH = partial(tabulate, tablefmt="orgtbl", headers=["row", "column"])
Middle
Backtrace
The backtrace algorithm traces a path through a minimum-edit-distance table to help us to optimally align substrings. How? Let's break that question up. First, how do you do it? We'll just use a greedy search that minimizes the cost. Starting at the last cell in the table (the minimum edit distance cell) we look at the cells directly above, directly to the left, and directly diagonal to the cell and move to the one of those three that has the lowest cost. We keep doing this until we reach the origin (0,0) cell and then we reverse the order of the cells we visited.
def backtrace(distances: numpy.ndarray) -> list:
"""finds the path for string alignment using backtrace
Args:
distances: array of mimimum edit distances
"""
# start at the bottom right cell
current_row, current_column = len(distances) - 1, len(distances[0]) - 1
path = [(current_row, current_column)]
while (current_row, current_column) != (0, 0):
one_row_back = current_row - 1
one_column_up = current_column - 1
edits = (
# insert
(distances[one_row_back, current_column], (one_row_back, current_column)),
# delete
(distances[current_row, one_column_up], (current_row, one_column_up)),
# substitute
(distances[one_row_back, one_column_up], (one_row_back, one_column_up))
)
minimum_edit_distance, cell_coordinates = min(edits)
path.append(cell_coordinates)
current_row, current_column = cell_coordinates
return list(reversed(path))
Simple Examples
- One Letter
We'll start with the simplest case, two strings with the same letter. Here's the distance table.
editor = MinimumEdits("a", "a") print(editor)
# a # 0 1 a 1 0 And here's the path through the table.
print(PATH(backtrace(editor.distance_table)))
row column 0 0 1 1 So, we start at the top-left and move to the bottom-right. Not too exciting…
Now let's try adding a second letter to the target word.
editor = MinimumEdits("a", "at") print(editor)
# a t # 0 1 2 a 1 0 1 path = backtrace(editor.distance_table) print(PATH(path))
row column 0 0 1 1 1 2 So we move from the top left then diagonally down and then laterally to the right. This gives us the first example of how the path is telling us to align the strings. Whenever the path moves horizontally (the row doesn't change) then that means you want to skip the character in the source.
Alignment
The rules for skipping characters as we move through the cells in the path are:
- If the current row is the same as the previous one, skip the character in the source, but add the character from the target.
- If the current column is the same as the previous one, skip the character in the target but add the character from the source.
def alignment(path: list, source: str, target: str,
empty_token: str="*") -> None:
"""Prints the alignment for the path
Args:
path: list of (row, column) tuples
source: the source string
target: the target string
empty_token: token to insert for skipped characters
"""
previous_row = previous_column = None
source_tokens = []
target_tokens = []
source = empty_token + source
target = empty_token + target
for current_row, current_column in path[1:]:
source_token = source[current_row] if current_row != previous_row else empty_token
target_token = target[current_column] if current_column != previous_column else empty_token
source_tokens.append(source_token)
target_tokens.append(target_token)
previous_row, previous_column = current_row, current_column
for tokens in (source_tokens, target_tokens):
print(f"|{'|'.join(tokens)}|")
return
Our alignment for the previous path looks like this.
alignment(path, "a", "at")
| a | * |
| a | t |
Where the * means skip that character. Okay, that might be obvious, but what if we have to skip the first letter?
editor = MinimumEdits("t", "at")
print(editor)
| # | a | t | |
|---|---|---|---|
| # | 0 | 1 | 2 |
| t | 1 | 2 | 1 |
path = backtrace(editor.distance_table)
print(PATH(path))
| row | column |
|---|---|
| 0 | 0 |
| 0 | 1 |
| 1 | 2 |
So in the first two cells the row doesn't change meaning that we skip the first letter in the source.
| a | t |
| * | t |
A Little More Interesting Example
Let's try a slightly more interesting example, aligning "drats" and "maths". First, the distance table.
SOURCE, TARGET = "drats", "maths"
editor = MinimumEdits(SOURCE, TARGET)
print(editor)
| # | m | a | t | h | s | |
|---|---|---|---|---|---|---|
| # | 0 | 1 | 2 | 3 | 4 | 5 |
| d | 1 | 2 | 3 | 4 | 5 | 6 |
| r | 2 | 3 | 4 | 5 | 6 | 7 |
| a | 3 | 4 | 3 | 4 | 5 | 6 |
| t | 4 | 5 | 4 | 3 | 4 | 5 |
| s | 5 | 6 | 5 | 4 | 5 | 4 |
Let's plot a heat map for it. If we plot the table as-is it ends up with the rows reversed, so we'll have to reverse the rows before plotting it.
reversed_table = editor.distance_frame.iloc[::-1]
plot = reversed_table.hvplot.heatmap(cmap=Plot.color_map).opts(
title="Minimum Edit Distances",
width=Plot.width, height=Plot.height, fontscale=Plot.fontscale
)
plot *= holoviews.Labels(plot)
outcome = Embed(plot=plot, file_name="drats_maths_distance_table")()
print(outcome)
Now we can take a look at the path.
path = backtrace(editor.distance_table)
print(PATH(path))
| row | column |
|---|---|
| 0 | 0 |
| 1 | 0 |
| 2 | 1 |
| 3 | 2 |
| 4 | 3 |
| 4 | 4 |
| 5 | 5 |
Now it's getting a little harder to see what's going on so let's plot the path along with the heatmap.
table = numpy.zeros(editor.distance_table.shape)
for row, column in path:
table[row, column] = 10
table = pandas.DataFrame(table, index=list("#drats"), columns=list("#maths"))
table = table.iloc[::-1]
path_plot = table.hvplot.heatmap(colorbar=False, cmap=Plot.path_color_map).opts(
title="Path For Alignment", width=1000, height=300)
distance_plot = reversed_table.hvplot.heatmap(cmap=Plot.color_map).opts(
title="Minimum Edit Distances", width=1000, height=300
)
plot = (path_plot + distance_plot).cols(1).opts(
width=800,
height=600,
fontscale=2,
)
outcome = Embed(plot=plot, file_name="drats_maths_alignment")()
print(outcome)
In the top plot the dark-blue rectangles are the ones chosen by the backtrace and the lower plot is a heatmap of the distances for each cell in the distance-table. You can sort of see that the path matches the cooler (smaller distance) cells in the distance heat map as you work from the top-left cell to the bottom-right cell (the minimum edit distance).
To interpret the path: where the column repeats you skip a character in the target and where the row repeats you skip a character in the source so our alignment looks like this.
alignment(path, SOURCE, TARGET)
| d | r | a | t | * | s |
| * | m | a | t | h | s |
Examples From Dan Jurasky
These are examples from Dan Jurasky's CS 124 lecture slides.
Nucleotides
Using the bokeh backend for heatmaps doesn't let you use column and index names that repeat, and I haven't figured out how to set x-ticks and y-ticks explicitly so I'll do it in matplotlib instead.
HEIGHT, WIDTH = 300, 1000
SOURCE, TARGET = "AGGCTATCACCTGACCTCCAGGCCGATGCCC", "TAGCTATCACGACCGCGGTCGATTTGCCCGAC"
editor = MinimumEdits(SOURCE, TARGET)
path = backtrace(editor.distance_table)
The plotting didn't work for this set so I'm not showing it (it scrambled the order and reduced the number of characters).
alignment(path, SOURCE, TARGET)
| * | A | G | G | C | T | A | T | C | A | C | C | T | G | A | C | C | T | C | C | A | G | G | * | C | C | G | A | T | * | * | G | C | C | C | * | * | * |
| T | A | G | * | C | T | A | T | C | A | C | * | * | G | A | C | C | G | C | * | * | G | G | T | C | * | G | A | T | T | T | G | C | C | C | G | A | C |
Intention and Execution
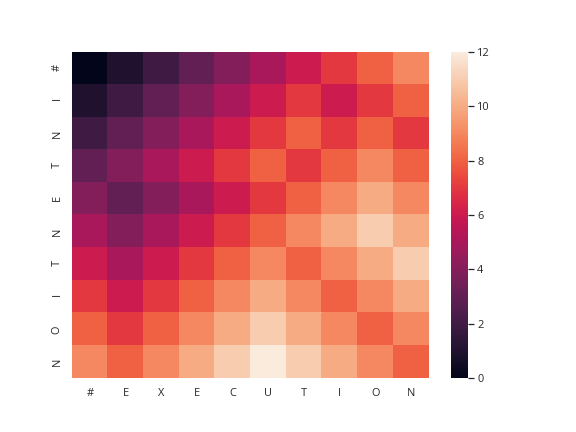
SOURCE, TARGET = "INTENTION", "EXECUTION"
editor = MinimumEdits(SOURCE, TARGET)
path = backtrace(editor.distance_table)
alignment(path, SOURCE, TARGET)
| I | N | T | E | * | * | * | N | T | I | O | N |
| * | * | * | E | X | E | C | U | T | I | O | N |
The output given by the presentation is
| I | N | T | E | * | N | T | I | O | N |
| * | E | X | E | C | U | T | I | O | N |
But I can't find anyplace where he documents how he derives these alignments so I don't know how to get this form.
figure, axis = pyplot.subplots()
grid = seaborn.heatmap(editor.distance_frame, ax=axis)
figure.savefig(FOLDER + "intention.png")
What About Sentences?
SOURCE = "he was big and bold and tall but old"
TARGET = "he is big i'm told but old"
editor = MinimumEdits(SOURCE, TARGET)
path = backtrace(editor.distance_table)
alignment(path, SOURCE, TARGET)
| h | e | w | a | s | b | i | g | a | n | d | b | o | l | d | a | n | d | t | a | l | l | b | u | t | o | l | d | ||||||||
| h | e | * | i | s | b | i | g | i | ' | m | t | o | l | d | * | * | * | * | * | * | * | * | * | b | u | t | o | l | d |
Sort of, but I'm sure that's not the right way to do it.
End
NLTK
The NLTK has a function that will get the path for us. It sort of hides the table from us (there's a private(ish) function that you can call to make the path if you have the table, otherwise they build the table and return the path). I couldn't find a direct link to it, but the it's in the metrics.distance module and is called edit_distance_align.
Let's see what it does with our last example.
nltk_path = edit_distance_align("drats", "maths")
print(f"|Ours| NLTK's|")
print(f"|-+-|")
for theirs, ours in zip(path, nltk_path):
print(f"|{ours}|{theirs}|")
| Ours | NLTK's |
|---|---|
| (0, 0) | (0, 0) |
| (1, 1) | (1, 0) |
| (2, 2) | (2, 1) |
| (3, 2) | (3, 2) |
| (4, 3) | (4, 3) |
| (5, 4) | (4, 4) |
| (5, 5) | (5, 5) |
So, you can see that ours doesn't really agree with theirs - which one of us is wrong?
help(edit_distance_align)
Help on function edit_distance_align in module nltk.metrics.distance:
edit_distance_align(s1, s2, substitution_cost=1)
Calculate the minimum Levenshtein edit-distance based alignment
mapping between two strings. The alignment finds the mapping
from string s1 to s2 that minimizes the edit distance cost.
For example, mapping "rain" to "shine" would involve 2
substitutions, 2 matches and an insertion resulting in
the following mapping:
[(0, 0), (1, 1), (2, 2), (3, 3), (4, 4), (4, 5)]
NB: (0, 0) is the start state without any letters associated
See more: https://web.stanford.edu/class/cs124/lec/med.pdf
In case of multiple valid minimum-distance alignments, the
backtrace has the following operation precedence:
1. Skip s1 character
2. Skip s2 character
3. Substitute s1 and s2 characters
The backtrace is carried out in reverse string order.
This function does not support transposition.
:param s1, s2: The strings to be aligned
:type s1: str
:type s2: str
:type substitution_cost: int
:rtype List[Tuple(int, int)]
Well, it looks like their substitution cost is 1 by default, not 2 like we're using. Take two.
nltk_align = partial(edit_distance_align, substitution_cost=2)
nltk_path = nltk_align("drats", "maths")
print(f"|Ours| NLTK's|")
print(f"|-+-|")
for ours, theirs in zip(path, nltk_path):
print(f"|{ours}|{theirs}|")
assert ours == theirs
| Ours | NLTK's |
|---|---|
| (0, 0) | (0, 0) |
| (1, 0) | (1, 0) |
| (2, 1) | (2, 1) |
| (3, 2) | (3, 2) |
| (4, 3) | (4, 3) |
| (4, 4) | (4, 4) |
| (5, 5) | (5, 5) |
So, if we're wrong, we're at least as wrong as NLTK is. Maybe.
Bundling It Up
Imports
# from pypi
import attr
# this repo
from neurotic.nlp.autocorrect.distance import MinimumEdits
The Aligner
@attr.s(auto_attribs=True)
class Aligner:
"""Create the alignment path
Args:
source: the source string to align
target: the target string to align
empty_token: character to use to fill in alignments
"""
source: str
target: str
empty_token: str="*"
_source_alignment: list=None
_target_alignment: list=None
_table: str=None
_editor: MinimumEdits=None
_path: list=None
The Editor
@property
def editor(self) -> MinimumEdits:
"""object to figure out the minimum edit distance"""
if self._editor is None:
self._editor = MinimumEdits(self.source, self.target)
return self._editor
The Alignment Path
@property
def path(self) -> list:
"""An optimal path through the distance table"""
if self._path is None:
distances = self.editor.distance_table
# start at the bottom right cell
current_row, current_column = (len(distances) - 1,
len(distances[0]) - 1)
path = [(current_row, current_column)]
while (current_row, current_column) != (0, 0):
one_row_back = current_row - 1
one_column_up = current_column - 1
edits = (
# insert
(distances[one_row_back, current_column], (one_row_back, current_column)),
# delete
(distances[current_row, one_column_up], (current_row, one_column_up)),
# substitute
(distances[one_row_back, one_column_up], (one_row_back, one_column_up))
)
minimum_edit_distance, cell_coordinates = min(edits)
path.append(cell_coordinates)
current_row, current_column = cell_coordinates
self._path = list(reversed(path))
return self._path
Source Alignment
@property
def source_alignment(self) -> list:
"""the aligned source tokens
Warning:
this doesn't create them, call the object to do that
"""
return self._source_alignment
Target Alignment
@property
def target_alignment(self) -> list:
"""The aligned target tokens
Warning:
this doesn't create them, the __call__ does
"""
return self._target_alignment
The Table
@property
def table(self) -> str:
"""The alignments as an orgtable"""
if self._table is None:
if self.source_alignment is None or self.target_alignment is None:
self()
self._table = (f"|{'|'.join(self.source_alignment)}|\n"
f"|{'|'.join(self.target_alignment)}|")
return self._table
The Call
def __call__(self) -> tuple:
"""Sets the source and target token alignments
Note:
as a side-effect also sets source_alignment and target_alignment
Returns:
tuple of source and target tokens after alignment
"""
previous_row = previous_column = None
source_tokens = []
target_tokens = []
source = self.empty_token + self.source
target = self.empty_token + self.target
for current_row, current_column in self.path[1:]:
source_token = (
source[current_row] if current_row != previous_row
else self.empty_token)
target_token = (
target[current_column] if current_column != previous_column
else self.empty_token)
source_tokens.append(source_token)
target_tokens.append(target_token)
previous_row, previous_column = current_row, current_column
self._source_alignment = source_tokens
self._target_alignment = target_tokens
return source_tokens, target_tokens
The String Representation
def __str__(self) -> str:
"""pass-through for the table"""
return self.table
Test It
from neurotic.nlp.autocorrect.alignment import Aligner
align = Aligner("source", "target")
print(align.editor)
| # | t | a | r | g | e | t | |
|---|---|---|---|---|---|---|---|
| # | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
| s | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| o | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| u | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| r | 4 | 5 | 6 | 5 | 6 | 7 | 8 |
| c | 5 | 6 | 7 | 6 | 7 | 8 | 9 |
| e | 6 | 7 | 8 | 7 | 8 | 7 | 8 |
print(PATH(align.path))
| row | column |
|---|---|
| 0 | 0 |
| 1 | 0 |
| 2 | 1 |
| 3 | 2 |
| 4 | 3 |
| 5 | 4 |
| 6 | 5 |
| 6 | 6 |
print(align())
(['s', 'o', 'u', 'r', 'c', 'e', '*'], ['*', 't', 'a', 'r', 'g', 'e', 't'])
print(align.table)
| s | o | u | r | c | e | * |
| * | t | a | r | g | e | t |
print(align)
| s | o | u | r | c | e | * |
| * | t | a | r | g | e | t |
align = Aligner("drafts", "maths")
nltk_path = nltk_align("drafts", "maths")
for ours, theirs in zip(align.path, nltk_path):
assert ours == theirs, f"{theirs}\t{ours}"
print(align)
| d | r | a | f | t | * | s |
| * | m | a | * | t | h | s |